shell scripts:
layersection.sh
Syntax
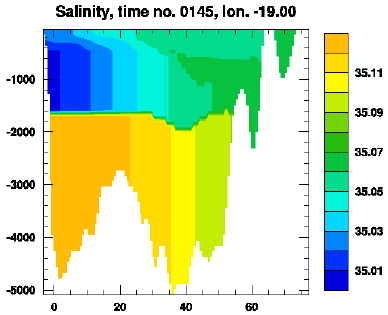
Example: Meridional cross-section of salinity
The figure below was made by issuing the command:
layersection.sh hycom_031_h.nc hycom_031_TS.nc thknss salin lat 62 145
layersection.sh: syntax
layersection.sh --help
layersection.sh / ncl-metno 1.2
>>>
>>>
>>> NOTE: This script assumes that the variable to depict has been
>>> stored with dimensions in the order x -y -layer(-time)
>>> or lon-lat-layer(-time)
>>> (w/ Fortran style sequence of dimensions).
>>>
>>> Syntax:
>>> =======
>>>
>>> layersection.sh (layer) <hfile> <varfile> <hname> <varname>\
>>> <dimname> <node> <time>
>>> where
>>> (layer) include an initial argument 'layer' if you want isopleths
>>> to be drawn for layer interfaces, ignore otherwise
>>> <hfile> name of netCDF file w/thickness results
>>> <varfile> name of netCDF file w/requested variable
>>> you may use '.' if both variables are on the same file
>>> <hname> name of thickness variable
>>> <varname> name of variable to depict
>>> NOTE! This script requires the existence of an
>>> attribute 'missing_value' to <varname>
>>> <dimname> name of crossection's horizontal dimension
>>> e.g., lat for a lat-z (meridional) crossection
>>> <node> node of non-depicted dimension
>>> if <dimname> is lat , this is the lon grid no.
>>> <time> time step no. to depict (use 0 for x-y-z & lon-lat-z fields)
>>> a negative <time> value is interpreted as a flag that
>>> stops the ncl script from being deleted
>>>
>>> The script will produce an eps-file and a png-file.
>>>
>>>
>>> User specifications:
>>> ====================
>>>
>>> By copying the default spec.s from
>>> /home/arnem/lib/ncl-metno/userdef.ncl
>>> to the directory where the command 'layersection.sh' is given,
>>> the user may specify
>>> * title
>>> * font
>>> * zooming
>>> * color map (palette)
>>> * no. of colors
>>> * plot size limits
>>> (look up, or copy, this file to edit your own 'userdef' file).
>>>
>>>
>>> Examples:
>>> =========
>>>
>>> layersection.sh hycom_expt007.nc . thknss salin lat 30 49
>>> will produce a lat-z crossection normal to longitude node no. 30,
>>> of the variable 'salin' on the file 'hycom_expt007.nc',
>>> w/ layers given by 'thknss' on the same file, add isopleths for
>>> layer interfaces, from timestep no. 49
>>> layersection.sh hycom_expt007.nc . thknss . lat 30 49
>>> same as above, but here, thknss is requested; this is a
>>> special case where the layer no. will be contoured
>>> (the thickess will correspond to the distance between layer
>>> interfaces), isopleths for layer interfaces will not be drawn
>>>
>>>
>>> Terminating.